 |
Timothy
0.9
Tissue Modelling Framework
|
 |
Timothy
0.9
Tissue Modelling Framework
|
contains functions which control current states and evolution of cells More...
#include <stdio.h>#include <stdlib.h>#include <mpi.h>#include <math.h>#include <inttypes.h>#include <sprng.h>#include "global.h"#include "inline.h"#include "fields.h"
Go to the source code of this file.
Functions | |
| void | cellsAllocate () |
| void | cellsCycleInit () |
| int | cellsRandomInit () |
| void | mitosis (int c) |
| void | markMiddleCancerCell () |
| void | cellsCleanup () |
| void | cellsDeath (int lnc_old) |
| void | updateCellCounters () |
| void | updateCellPositions () |
| int | updateCellCycles () |
| void | additionalScalarField () |
| void | updateCellStates () |
Variables | |
| unsigned char * | celld |
contains functions which control current states and evolution of cells
Definition in file cells.c.
| void additionalScalarField | ( | ) |
This function fills the scalarOutput field of each cell. It can be modified to output any float scalars that user would like to analyze or visualize after simulation. This field is printed to the output VTK files.
Definition at line 704 of file cells.c.
References cells, cellData::density, lnc, and cellData::scalarField.

| void cellsAllocate | ( | ) |
This function allocates tables responsible for carrying informations about cells, their current state and evolution.
Definition at line 66 of file cells.c.
References cellFields, cells, csize, localID, maxCells, maxCellsPerProc, MPIsize, NFIELDS, nx, sdim, stopRun(), tlnc, and velocity.

| void cellsCleanup | ( | ) |
| void cellsCycleInit | ( | ) |
| void cellsDeath | ( | int | lnc_old | ) |
| int cellsRandomInit | ( | ) |
This function initializes cell data. Locations of cells in space are generated randomly.
Definition at line 132 of file cells.c.
References cellData::age, cells, csize, cellData::death, cellData::density, cellData::g1, g1, cellData::g2, g2, cellData::gid, cellData::h, h, h2, h3, h4, cellData::halo, lnc, localID, cellData::m, m, maxCells, maxCellsPerProc, MPIrank, nc, nx, cellData::phase, cellData::phasetime, rng, cellData::s, s, sdim, simTime, cellData::size, stream, cellData::tumor, cellData::v, v, cellData::x, cellData::y, cellData::young, and cellData::z.

| void markMiddleCancerCell | ( | ) |
This function finds locates cell closest to the center of mass of the system and marks this cell as a cancer cell.
Definition at line 374 of file cells.c.
References cancer, cells, lcnc, lg0nc, lg1nc, lnc, MPIrank, nc, cellData::phase, cellData::tumor, x, cellData::x, cellData::y, and cellData::z.

| void mitosis | ( | int | c | ) |
This function implements mitosis of cells.
Definition at line 266 of file cells.c.
References cellData::age, cells, cg1, cg2, cm, cs, cellData::death, cellData::density, cellData::g1, g1, cellData::g2, g2, cellData::gid, cellData::h, h, cellData::halo, lcnc, lg1nc, lnc, localID, cellData::m, m, maxCellsPerProc, mitrand, MPIrank, cellData::phase, cellData::phasetime, cellData::s, s, sdim, cellData::size, stopRun(), stream, cellData::tumor, cellData::v, v, velocity, x, cellData::x, doubleVector3d::x, cellData::y, doubleVector3d::y, cellData::young, cellData::z, and doubleVector3d::z.


| void updateCellCounters | ( | ) |
This function updates cell counters.
Definition at line 493 of file cells.c.
References lnc, localCellCount, numberOfCounts, tlnc, and totalCellCount.

| int updateCellCycles | ( | ) |
This function updates cells' cycle phases.
Definition at line 537 of file cells.c.
References celld, cellFields, cells, csize, densityCriticalLevel1, densityCriticalLevel2, fieldCriticalLevel1, fieldCriticalLevel2, cellData::g1, g1, g2, gfDt, gfields, lg0nc, lg1nc, lg2nc, lmnc, lnc, lnnc, lsnc, m, mitosis(), nc, OXYG, oxygen, cellData::phase, cellData::phasetime, rd, rsum, s, simStart, cellData::size, and stream.


| void updateCellPositions | ( | ) |
This function updates cells' positions.
Definition at line 504 of file cells.c.
References cells, csize, lnc, statisticsData::mindist, MPIrank, nc, simStart, statistics, statOutStep, step, velocity, cellData::x, doubleVector3d::x, cellData::y, doubleVector3d::y, cellData::z, and doubleVector3d::z.

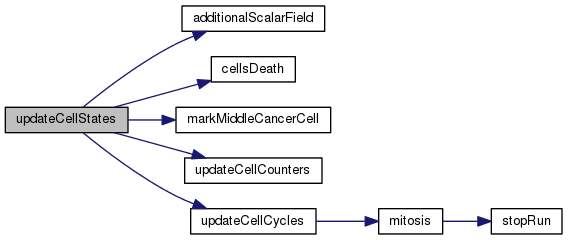
| void updateCellStates | ( | ) |
This function drives the whole cell cycle update.
Definition at line 718 of file cells.c.
References additionalScalarField(), cancer, celld, cellsDeath(), lnc, markMiddleCancerCell(), nc, nhs, rsum, tgs, updateCellCounters(), and updateCellCycles().